4 Chapter 14
This is the code for Chapter 14
4.1 Program 14.1
ranks <- nhefs %>%
mutate(
rank = min_rank(desc(wt82_71)),
lbl = if_else(
rank <= 3 | rank >= (max(rank, na.rm = TRUE) - 2),
round(wt82_71, 1),
NA_real_
)
) %>%
select(seqn, rank, lbl, wt82_71)
ranks %>%
select(-lbl) %>%
top_n(5, wt82_71)## # A tibble: 5 x 3
## seqn rank wt82_71
## <dbl> <int> <dbl>
## 1 1769 3 37.7
## 2 5415 5 34.0
## 3 6928 2 47.5
## 4 22342 4 37.0
## 5 23522 1 48.5## # A tibble: 5 x 3
## seqn rank wt82_71
## <dbl> <int> <dbl>
## 1 5412 1563 -29.0
## 2 13593 1565 -30.5
## 3 21897 1562 -26.0
## 4 23321 1566 -41.3
## 5 24363 1564 -30.1ranks %>%
ggplot(aes(y = rank, x = wt82_71)) +
geom_vline(xintercept = 0, col = "grey90", size = 1.3) +
geom_point(col = "#0072B2", size = 1, alpha = .9) +
ggrepel::geom_text_repel(
aes(label = lbl),
size = 4,
point.padding = 0.1,
box.padding = .6,
force = 1.,
min.segment.length = 0,
seed = 777
) +
theme_minimal(14) +
expand_limits(y = c(-200, 1700)) +
xlab("change in weight")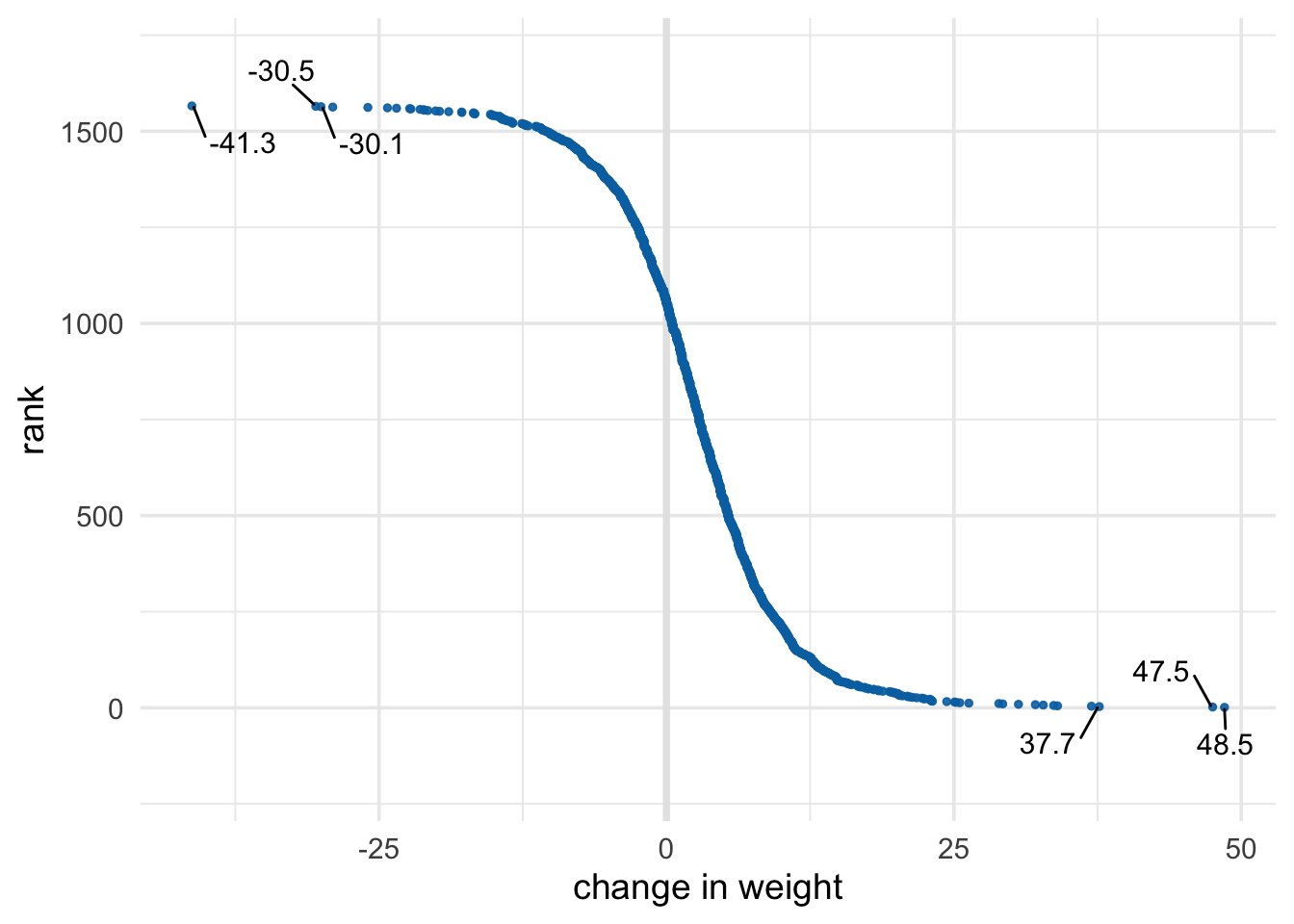
4.2 Program 14.2
# using complete data set
nhefs_censored <- nhefs %>%
drop_na(qsmk, sex, race, age, school, smokeintensity, smokeyrs, exercise,
active, wt71)
# compute unstabilized inverse probability of censoring weights
cwts_model <- glm(
censored ~ qsmk + sex + race + age + I(age^2) + education +
smokeintensity + I(smokeintensity^2) +
smokeyrs + I(smokeyrs^2) + exercise + active +
wt71 + I(wt71^2),
data = nhefs_censored, family = binomial()
)
nhefs_censored <- cwts_model %>%
augment(type.predict = "response", data = nhefs_censored) %>%
mutate(cwts = 1 / ifelse(censored == 0, 1 - .fitted, .fitted))
# compute all values of h(psi)
compute_h_psi <- function(psi) {
df <- nhefs_censored %>%
mutate(h_psi = wt82_71 - psi * qsmk) %>%
# gee doesn't like missing values
drop_na(h_psi)
geeglm(
qsmk ~ h_psi + sex + race + age + I(age^2) + education +
smokeintensity + I(smokeintensity^2) +
smokeyrs + I(smokeyrs^2) + exercise + active +
wt71 + I(wt71^2),
data = df,
family = binomial(),
std.err = "san.se",
weights = cwts,
id = id,
corstr = "independence"
) %>%
tidy() %>%
filter(term == "h_psi") %>%
mutate(psi = psi) %>%
select(psi, estimate, p.value)
}
compute_h_psi(3.446)## # A tibble: 1 x 3
## psi estimate p.value
## <dbl> <dbl> <dbl>
## 1 3.45 -0.00000190 1.000## # A tibble: 31 x 3
## psi estimate p.value
## <dbl> <dbl> <dbl>
## 1 3.4 0.000862 0.922
## 2 3.5 -0.00102 0.909
## 3 3.3 0.00274 0.755
## 4 3.6 -0.00290 0.744
## 5 3.2 0.00461 0.598
## 6 3.7 -0.00480 0.592
## 7 3.1 0.00647 0.457
## 8 3.8 -0.00670 0.457
## 9 3 0.00833 0.337
## 10 3.9 -0.00860 0.342
## # … with 21 more rowspsi_est <- psi_search %>%
arrange(abs(estimate)) %>%
slice(1) %>%
select(-estimate, -p.value)
# get minimum and maximum values that have p >= .05 for confidence intervals
psi_conf_int <- psi_search %>%
filter(p.value >= .05) %>%
slice(c(1, n())) %>%
mutate(type = c("conf.low", "conf.high")) %>%
select(type, psi) %>%
spread(type, psi)
bind_cols(psi_est, psi_conf_int)## # A tibble: 1 x 3
## psi conf.high conf.low
## <dbl> <dbl> <dbl>
## 1 3.4 4.4 2.6psi_search %>%
ggplot(aes(x = psi, y = estimate)) +
geom_hline(yintercept = 0, col = "grey85", size = 1.3) +
geom_line(col = "#0072B2", size = 1.2) +
geom_point(shape = 21, col = "white", fill = "#0072B2", size = 2.5) +
theme_minimal(14)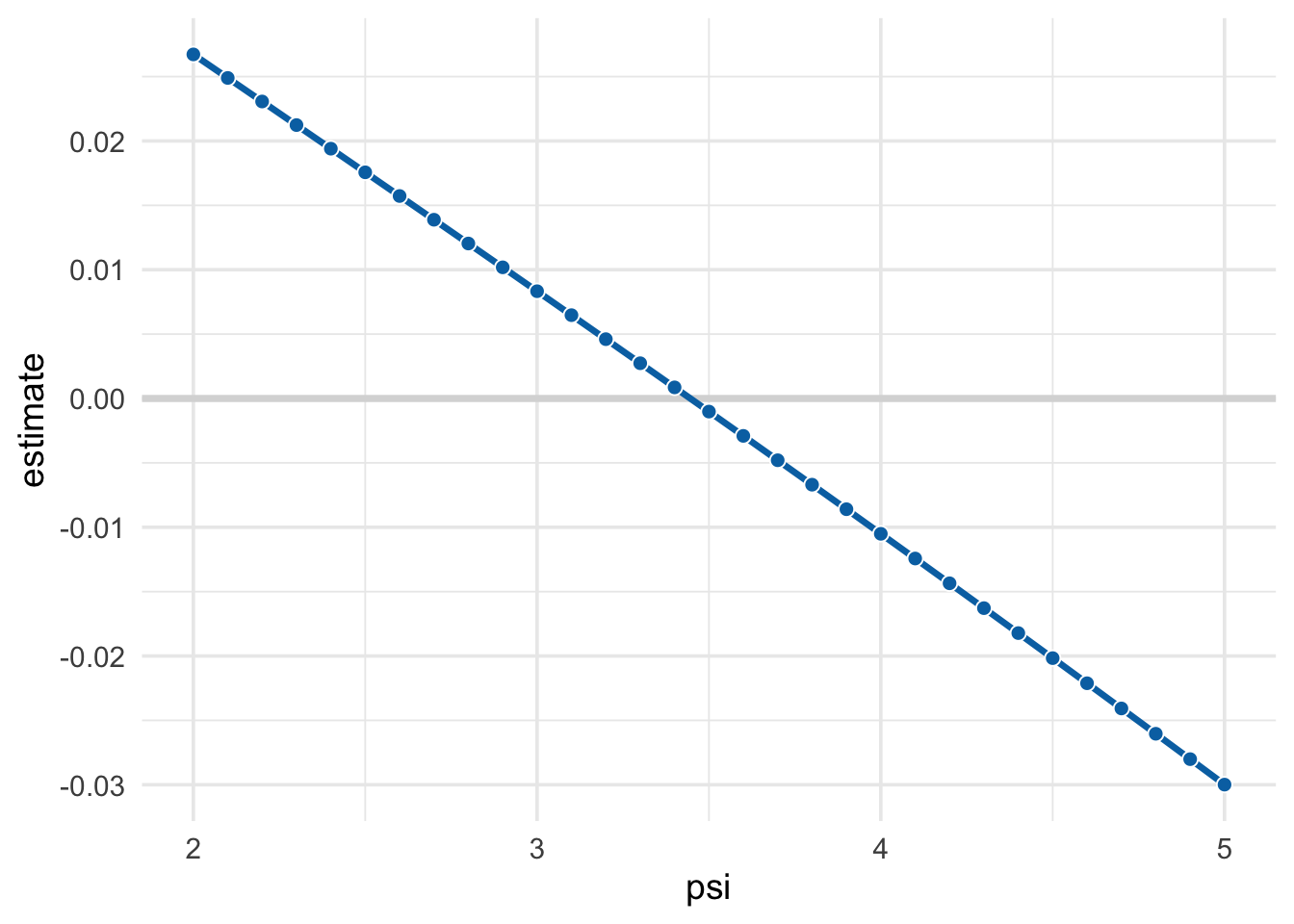
psi_formula <- function(weights, outcome, treatment, treatment_pred) {
numerator <- weights * outcome * (treatment - treatment_pred)
denominator <- sum(weights * treatment * (treatment - treatment_pred), na.rm = TRUE)
sum(numerator / denominator, na.rm = TRUE)
}
estimate_psi <- function(.data) {
glm(
qsmk ~ sex + race + age + I(age^2) + education +
smokeintensity + I(smokeintensity^2) +
smokeyrs + I(smokeyrs^2) + exercise + active +
wt71 + I(wt71^2),
data = .data,
family = binomial(),
weights = cwts,
) %>%
augment(data = .data, type.predict = "response") %>%
summarize(
psi = psi_formula(
weights = cwts,
outcome = wt82_71,
treatment = qsmk,
treatment_pred = .fitted
)
)
}
nhefs_censored %>%
select(-.fitted:-.cooksd) %>%
filter(censored == 0) %>%
estimate_psi()## # A tibble: 1 x 1
## psi
## <dbl>
## 1 3.45bootstrap_psi <- function(data, indices) {
estimate_psi(data[indices, ]) %>% pull(psi)
}
bootstrapped_psis <- nhefs_censored %>%
select(-.fitted:-.cooksd) %>%
filter(censored == 0) %>%
boot(bootstrap_psi, R = 2000)
bootstrapped_psis %>%
tidy(conf.int = TRUE, conf.method = "bca")## # A tibble: 1 x 5
## statistic bias std.error conf.low conf.high
## <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 3.45 -0.00202 0.473 2.51 4.384.3 Program 14.3
estimate_psi2 <- function(.data) {
glm(
qsmk ~ sex + race + age + I(age^2) + education +
smokeintensity + I(smokeintensity^2) +
smokeyrs + I(smokeyrs^2) + exercise + active +
wt71 + I(wt71^2),
data = .data,
family = binomial(),
weights = cwts,
) %>%
augment(data = .data, type.predict = "response") %>%
psi_formula2()
}
solve_matrix <- function(.data, .names = c("psi1", "psi2")) {
cells <- .data %>%
summarise(
a1 = sum(qsmk * diff),
a2 = sum(qsmk * smokeintensity * diff),
a3 = sum(qsmk * smokeintensity * diff),
a4 = sum(qsmk * smokeintensity * smokeintensity * diff),
b1 = sum(wt82_71 * diff),
b2 = sum(wt82_71 * smokeintensity * diff)
)
a <- cells %>%
select(a1:a4) %>%
unlist() %>%
matrix(2, 2)
b <- cells %>%
select(b1:b2) %>%
unlist() %>%
matrix(2, 1)
solve(a, b) %>%
t() %>%
as_tibble(.name_repair = "minimal") %>%
set_names(.names)
}
psi_formula2 <- function(.data) {
.data %>%
mutate(diff = (qsmk - .fitted) * cwts) %>%
drop_na(wt82_71) %>%
solve_matrix()
}
nhefs_censored %>%
select(-.fitted:-.cooksd) %>%
filter(censored == 0) %>%
estimate_psi2()## # A tibble: 1 x 2
## psi1 psi2
## <dbl> <dbl>
## 1 2.86 0.0300