2 Chapter 12
This is the code for Chapter 12
2.1 Program 12.1
## # A tibble: 1,629 x 64
## seqn qsmk death yrdth modth dadth sbp dbp sex age race income
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 233 0 0 NA NA NA 175 96 0 42 1 19
## 2 235 0 0 NA NA NA 123 80 0 36 0 18
## 3 244 0 0 NA NA NA 115 75 1 56 1 15
## 4 245 0 1 85 2 14 148 78 0 68 1 15
## 5 252 0 0 NA NA NA 118 77 0 40 0 18
## 6 257 0 0 NA NA NA 141 83 1 43 1 11
## 7 262 0 0 NA NA NA 132 69 1 56 0 19
## 8 266 0 0 NA NA NA 100 53 1 29 0 22
## 9 419 0 1 84 10 13 163 79 0 51 0 18
## 10 420 0 1 86 10 17 184 106 0 43 0 16
## # … with 1,619 more rows, and 52 more variables: marital <dbl>,
## # school <dbl>, education <dbl>, ht <dbl>, wt71 <dbl>, wt82 <dbl>,
## # wt82_71 <dbl>, birthplace <dbl>, smokeintensity <dbl>,
## # smkintensity82_71 <dbl>, smokeyrs <dbl>, asthma <dbl>, bronch <dbl>,
## # tb <dbl>, hf <dbl>, hbp <dbl>, pepticulcer <dbl>, colitis <dbl>,
## # hepatitis <dbl>, chroniccough <dbl>, hayfever <dbl>, diabetes <dbl>,
## # polio <dbl>, tumor <dbl>, nervousbreak <dbl>, alcoholpy <dbl>,
## # alcoholfreq <dbl>, alcoholtype <dbl>, alcoholhowmuch <dbl>,
## # pica <dbl>, headache <dbl>, otherpain <dbl>, weakheart <dbl>,
## # allergies <dbl>, nerves <dbl>, lackpep <dbl>, hbpmed <dbl>,
## # boweltrouble <dbl>, wtloss <dbl>, infection <dbl>, active <dbl>,
## # exercise <dbl>, birthcontrol <dbl>, pregnancies <dbl>,
## # cholesterol <dbl>, hightax82 <dbl>, price71 <dbl>, price82 <dbl>,
## # tax71 <dbl>, tax82 <dbl>, price71_82 <dbl>, tax71_82 <dbl>nhefs <- nhefs %>%
# add id and censored indicator
# recode age > 50 and years of school to categories
mutate(
id = 1:n(),
censored = ifelse(is.na(wt82), 1, 0),
older = case_when(is.na(age) ~ NA_real_,
age > 50 ~ 1,
TRUE ~ 0),
education = case_when(school < 9 ~ 1,
school < 12 ~ 2,
school == 12 ~ 3,
school < 16 ~ 4,
TRUE ~ 5)
) %>%
# change categorical to factors
mutate_at(vars(sex, race, education, exercise, active), factor)
# restrict to complete cases
nhefs_complete <- nhefs %>%
drop_na(qsmk, sex, race, age, school, smokeintensity, smokeyrs, exercise,
active, wt71, wt82, wt82_71, censored)
nhefs_complete %>%
filter(censored == 0) %>%
group_by(qsmk) %>%
summarize(mean_weight_change = mean(wt82_71), sd = sd(wt82_71)) %>%
knitr::kable(digits = 2)| qsmk | mean_weight_change | sd |
|---|---|---|
| 0 | 1.98 | 7.45 |
| 1 | 4.53 | 8.75 |
# clean up the data for the table
fct_yesno <- function(x) {
factor(x, labels = c("No", "Yes"))
}
tbl1_data <- nhefs_complete %>%
filter(censored == 0) %>%
mutate(university = fct_yesno(ifelse(education == 5, 1, 0)),
no_exercise = fct_yesno(ifelse(exercise == 2, 1, 0)),
inactive = fct_yesno(ifelse(active == 2, 1, 0)),
qsmk = factor(qsmk, levels = 1:0, c("Ceased Smoking", "Continued Smoking")),
sex = factor(sex, levels = 1:0, labels = c("Female", "Male")),
race = factor(race, levels = 1:0, labels = c("Other", "White"))) %>%
select(qsmk, age, sex, race, university, wt71, smokeintensity, smokeyrs, no_exercise,
inactive) %>%
rename("Smoking Cessation" = "qsmk",
"Age" = "age",
"Sex" = "sex",
"Race" = "race",
"University education" = "university",
"Weight, kg" = "wt71",
"Cigarettes/day" = "smokeintensity",
"Years smoking" = "smokeyrs",
"Little or no exercise" = "no_exercise",
"Inactive daily life" = "inactive")
kableone <- function(x, ...) {
capture.output(x <- print(x))
knitr::kable(x, ...)
}
tbl1_data %>%
CreateTableOne(vars = select(tbl1_data, -`Smoking Cessation`) %>% names,
strata = "Smoking Cessation", data = ., test = FALSE) %>%
kableone()| Ceased Smoking | Continued Smoking | |
|---|---|---|
| n | 403 | 1163 |
| Age (mean (sd)) | 46.17 (12.21) | 42.79 (11.79) |
| Sex = Male (%) | 220 (54.6) | 542 (46.6) |
| Race = White (%) | 367 (91.1) | 993 (85.4) |
| University education = Yes (%) | 62 (15.4) | 115 ( 9.9) |
| Weight, kg (mean (sd)) | 72.35 (15.63) | 70.30 (15.18) |
| Cigarettes/day (mean (sd)) | 18.60 (12.40) | 21.19 (11.48) |
| Years smoking (mean (sd)) | 26.03 (12.74) | 24.09 (11.71) |
| Little or no exercise = Yes (%) | 164 (40.7) | 441 (37.9) |
| Inactive daily life = Yes (%) | 45 (11.2) | 104 ( 8.9) |
2.2 Program 12.2
# Estimation of IP weights via a logistic model
propensity <- glm(qsmk ~ sex + race + age + I(age^2) + education +
smokeintensity + I(smokeintensity^2) +
smokeyrs + I(smokeyrs^2) + exercise + active +
wt71 + I(wt71^2),
family = binomial(), data = nhefs_complete)
propensity %>%
tidy(conf.int = TRUE, exponentiate = TRUE) %>%
select(-statistic, -p.value) %>%
knitr::kable(digits = 2)| term | estimate | std.error | conf.low | conf.high |
|---|---|---|---|---|
| (Intercept) | 0.11 | 1.38 | 0.01 | 1.56 |
| sex1 | 0.59 | 0.15 | 0.44 | 0.80 |
| race1 | 0.43 | 0.21 | 0.28 | 0.64 |
| age | 1.13 | 0.05 | 1.02 | 1.25 |
| I(age^2) | 1.00 | 0.00 | 1.00 | 1.00 |
| education2 | 0.97 | 0.20 | 0.66 | 1.43 |
| education3 | 1.09 | 0.18 | 0.77 | 1.55 |
| education4 | 1.07 | 0.27 | 0.62 | 1.81 |
| education5 | 1.61 | 0.23 | 1.03 | 2.51 |
| smokeintensity | 0.93 | 0.02 | 0.90 | 0.95 |
| I(smokeintensity^2) | 1.00 | 0.00 | 1.00 | 1.00 |
| smokeyrs | 0.93 | 0.03 | 0.88 | 0.98 |
| I(smokeyrs^2) | 1.00 | 0.00 | 1.00 | 1.00 |
| exercise1 | 1.43 | 0.18 | 1.01 | 2.04 |
| exercise2 | 1.49 | 0.19 | 1.03 | 2.15 |
| active1 | 1.03 | 0.13 | 0.80 | 1.34 |
| active2 | 1.19 | 0.21 | 0.78 | 1.81 |
| wt71 | 0.98 | 0.03 | 0.94 | 1.04 |
| I(wt71^2) | 1.00 | 0.00 | 1.00 | 1.00 |
nhefs_complete <- propensity %>%
augment(type.predict = "response", data = nhefs_complete) %>%
mutate(wts = 1 / ifelse(qsmk == 0, 1 - .fitted, .fitted))
nhefs_complete %>%
summarize(mean_wt = mean(wts), sd_wts = sd(wts))## # A tibble: 1 x 2
## mean_wt sd_wts
## <dbl> <dbl>
## 1 2.00 1.47ggplot(nhefs_complete, aes(wts)) +
geom_density(col = "#E69F00", fill = "#E69F0095") +
scale_x_log10() +
theme_minimal(base_size = 14) +
xlab("log10(Weights)")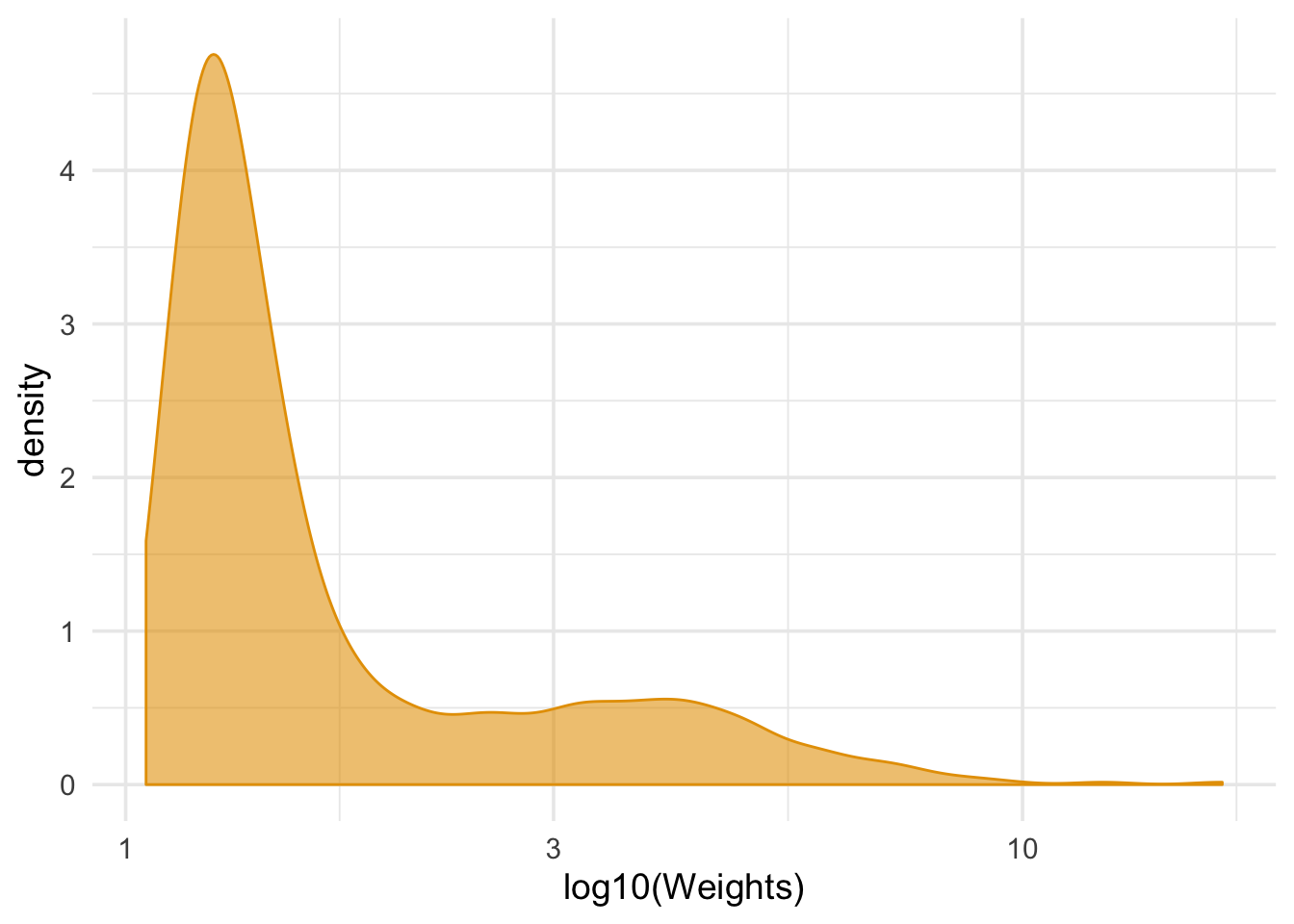
tidy_est_cis <- function(.df, .type) {
.df %>%
mutate(type = .type) %>%
filter(term == "qsmk") %>%
select(type, estimate, conf.low, conf.high)
}
# standard error a little too small
ols_cis <- lm(wt82_71 ~ qsmk, data = nhefs_complete, weights = wts) %>%
tidy(conf.int = TRUE) %>%
tidy_est_cis("ols")
ols_cis## # A tibble: 1 x 4
## type estimate conf.low conf.high
## <chr> <dbl> <dbl> <dbl>
## 1 ols 3.44 2.64 4.24library(geepack)
gee_model <- geeglm(
wt82_71 ~ qsmk,
data = nhefs_complete,
std.err = "san.se", # default robust SE
weights = wts, # inverse probability weights
id = id,
corstr = "independence" # independent correlation structure
)
gee_model_cis <- tidy(gee_model, conf.int = TRUE) %>%
tidy_est_cis("gee")
gee_model_cis## # A tibble: 1 x 4
## type estimate conf.low conf.high
## <chr> <dbl> <dbl> <dbl>
## 1 gee 3.44 2.41 4.47# easy robust SEs
library(estimatr)
robust_lm_model_cis <- lm_robust(
wt82_71 ~ qsmk, data = nhefs_complete,
weights = wts
) %>%
tidy() %>%
tidy_est_cis("robust ols")
robust_lm_model_cis## type estimate conf.low conf.high
## 1 robust ols 3.440535 2.407886 4.473185model_nhefs <- function(data, indices) {
# need to bootstrap the entire fitting process, including IPWs
df <- data[indices, ]
propensity <- glm(qsmk ~ sex + race + age + I(age^2) + education +
smokeintensity + I(smokeintensity^2) +
smokeyrs + I(smokeyrs^2) + exercise + active +
wt71 + I(wt71^2),
family = binomial(), data = df)
df <- propensity %>%
augment(type.predict = "response", data = df) %>%
mutate(wts = 1 / ifelse(qsmk == 0, 1 - .fitted, .fitted))
lm(wt82_71 ~ qsmk, data = df, weights = wts) %>%
tidy() %>%
filter(term == "qsmk") %>%
pull(estimate)
}library(boot)
set.seed(1234)
bootstrap_estimates <- nhefs_complete %>%
select(-.fitted:-wts) %>%
boot(model_nhefs, R = 2000)
bootstrap_cis <- bootstrap_estimates %>%
tidy(conf.int = TRUE, conf.method = "bca") %>%
mutate(type = "bootstrap") %>%
select(type, estimate = statistic, conf.low, conf.high)
bootstrap_cis## # A tibble: 1 x 4
## type estimate conf.low conf.high
## <chr> <dbl> <dbl> <dbl>
## 1 bootstrap 3.44 2.47 4.43bind_rows(ols_cis, gee_model_cis, robust_lm_model_cis, bootstrap_cis) %>%
mutate(width = conf.high - conf.low) %>%
arrange(width) %>%
mutate(type = fct_inorder(type)) %>%
ggplot(aes(x = type, y = estimate, ymin = conf.low, ymax = conf.high)) +
geom_pointrange(color = "#0172B1", size = 1, fatten = 3) +
coord_flip() +
theme_minimal(base_size = 14)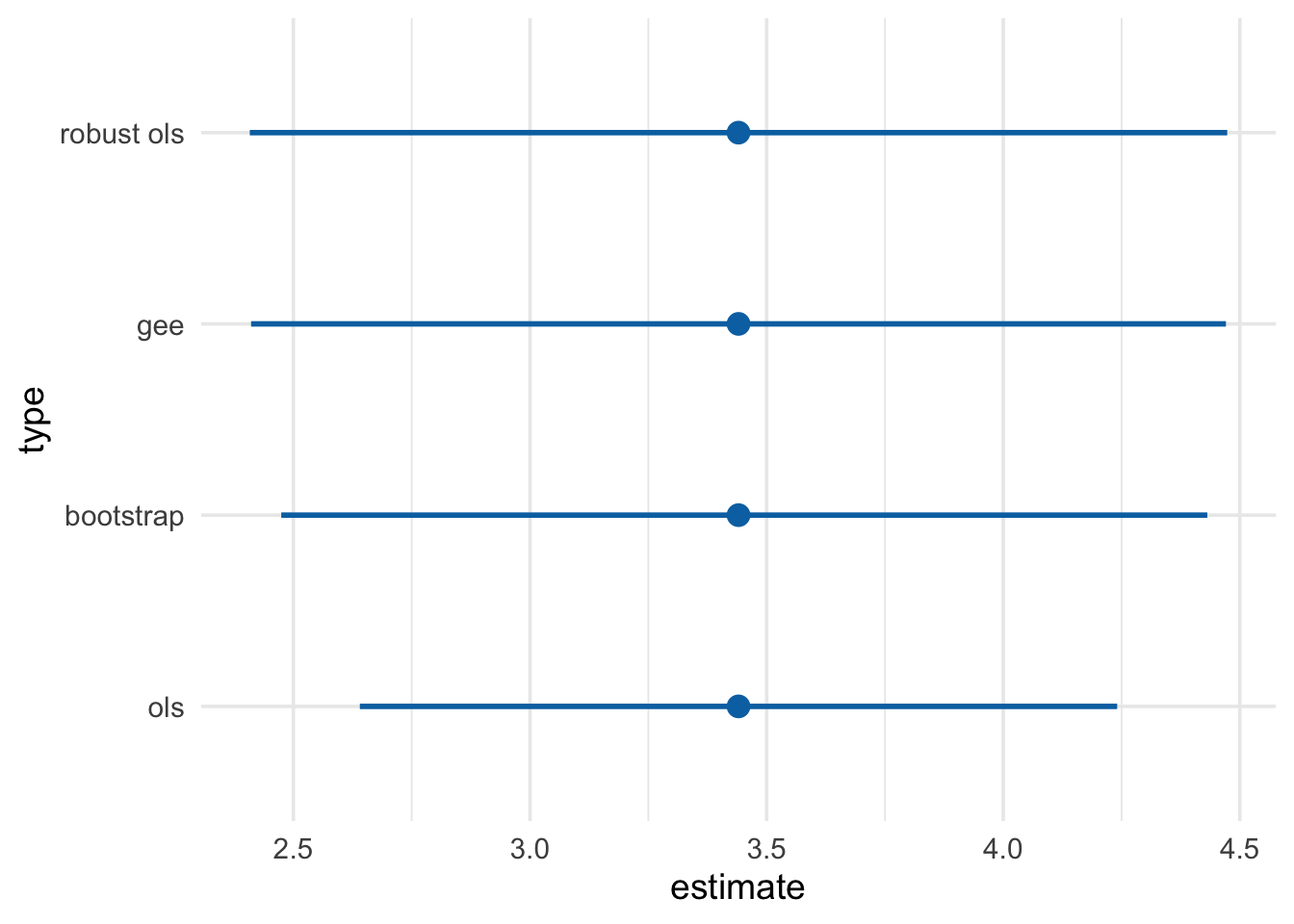
2.3 Program 12.3
numerator <- glm(qsmk ~ 1, data = nhefs_complete, family = binomial())
nhefs_complete <- numerator %>%
augment(type.predict = "response", data = nhefs_complete) %>%
mutate(numerator = ifelse(qsmk == 0, 1 - .fitted, .fitted)) %>%
select(id, numerator) %>%
left_join(nhefs_complete, by = "id") %>%
mutate(swts = numerator / ifelse(qsmk == 0, 1 - .fitted, .fitted))
nhefs_complete %>%
summarize(mean_wt = mean(swts), sd_wts = sd(swts))## # A tibble: 1 x 2
## mean_wt sd_wts
## <dbl> <dbl>
## 1 0.999 0.288ggplot(nhefs_complete, aes(swts)) +
geom_density(col = "#E69F00", fill = "#E69F0095") +
scale_x_log10() +
theme_minimal(base_size = 14) +
xlab("log10(Stabilized Weights)")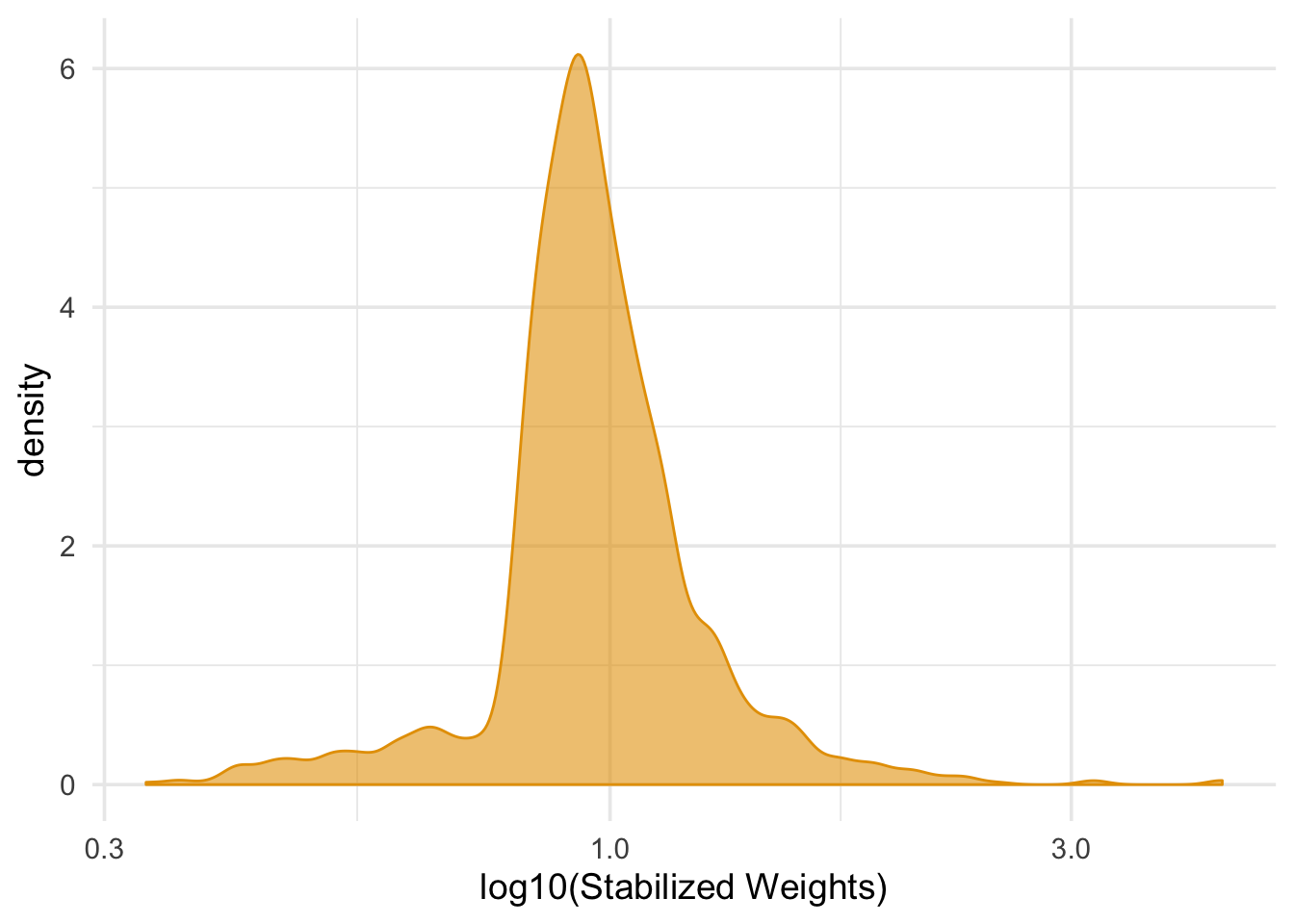
## term estimate std.error statistic p.value conf.low conf.high
## 1 (Intercept) 1.779978 0.2248362 7.916778 4.581734e-15 1.338966 2.220990
## 2 qsmk 3.440535 0.5264638 6.535179 8.573524e-11 2.407886 4.473185
## df outcome
## 1 1564 wt82_71
## 2 1564 wt82_712.4 Program 12.4
nhefs_light_smokers <- nhefs %>%
drop_na(qsmk, sex, race, age, school, smokeintensity, smokeyrs, exercise,
active, wt71, wt82, wt82_71, censored) %>%
filter(smokeintensity <= 25)
nhefs_light_smokers## # A tibble: 1,162 x 67
## seqn qsmk death yrdth modth dadth sbp dbp sex age race income
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <fct> <dbl> <fct> <dbl>
## 1 235 0 0 NA NA NA 123 80 0 36 0 18
## 2 244 0 0 NA NA NA 115 75 1 56 1 15
## 3 245 0 1 85 2 14 148 78 0 68 1 15
## 4 252 0 0 NA NA NA 118 77 0 40 0 18
## 5 257 0 0 NA NA NA 141 83 1 43 1 11
## 6 262 0 0 NA NA NA 132 69 1 56 0 19
## 7 266 0 0 NA NA NA 100 53 1 29 0 22
## 8 419 0 1 84 10 13 163 79 0 51 0 18
## 9 420 0 1 86 10 17 184 106 0 43 0 16
## 10 434 0 0 NA NA NA 127 80 1 54 0 16
## # … with 1,152 more rows, and 55 more variables: marital <dbl>,
## # school <dbl>, education <fct>, ht <dbl>, wt71 <dbl>, wt82 <dbl>,
## # wt82_71 <dbl>, birthplace <dbl>, smokeintensity <dbl>,
## # smkintensity82_71 <dbl>, smokeyrs <dbl>, asthma <dbl>, bronch <dbl>,
## # tb <dbl>, hf <dbl>, hbp <dbl>, pepticulcer <dbl>, colitis <dbl>,
## # hepatitis <dbl>, chroniccough <dbl>, hayfever <dbl>, diabetes <dbl>,
## # polio <dbl>, tumor <dbl>, nervousbreak <dbl>, alcoholpy <dbl>,
## # alcoholfreq <dbl>, alcoholtype <dbl>, alcoholhowmuch <dbl>,
## # pica <dbl>, headache <dbl>, otherpain <dbl>, weakheart <dbl>,
## # allergies <dbl>, nerves <dbl>, lackpep <dbl>, hbpmed <dbl>,
## # boweltrouble <dbl>, wtloss <dbl>, infection <dbl>, active <fct>,
## # exercise <fct>, birthcontrol <dbl>, pregnancies <dbl>,
## # cholesterol <dbl>, hightax82 <dbl>, price71 <dbl>, price82 <dbl>,
## # tax71 <dbl>, tax82 <dbl>, price71_82 <dbl>, tax71_82 <dbl>, id <int>,
## # censored <dbl>, older <dbl>denominator_model <- lm(smkintensity82_71 ~ sex + race + age + I(age^2) + education +
smokeintensity + I(smokeintensity^2) +
smokeyrs + I(smokeyrs^2) + exercise + active +
wt71 + I(wt71^2), data = nhefs_light_smokers)
denominators <- denominator_model %>%
augment(data = nhefs_light_smokers) %>%
mutate(denominator = dnorm(smkintensity82_71, .fitted, mean(.sigma, na.rm = TRUE))) %>%
select(id, denominator)
numerator_model <- lm(smkintensity82_71 ~ 1, data = nhefs_light_smokers)
numerators <- numerator_model %>%
augment(data = nhefs_light_smokers) %>%
mutate(numerator = dnorm(smkintensity82_71, .fitted, mean(.sigma, na.rm = TRUE))) %>%
select(id, numerator)
nhefs_light_smokers <- nhefs_light_smokers %>%
left_join(numerators, by = "id") %>%
left_join(denominators, by = "id") %>%
mutate(swts = numerator / denominator)
ggplot(nhefs_light_smokers, aes(swts)) +
geom_density(col = "#E69F00", fill = "#E69F0095") +
scale_x_log10() +
theme_minimal(base_size = 14) +
xlab("log10(Stabilized Weights)")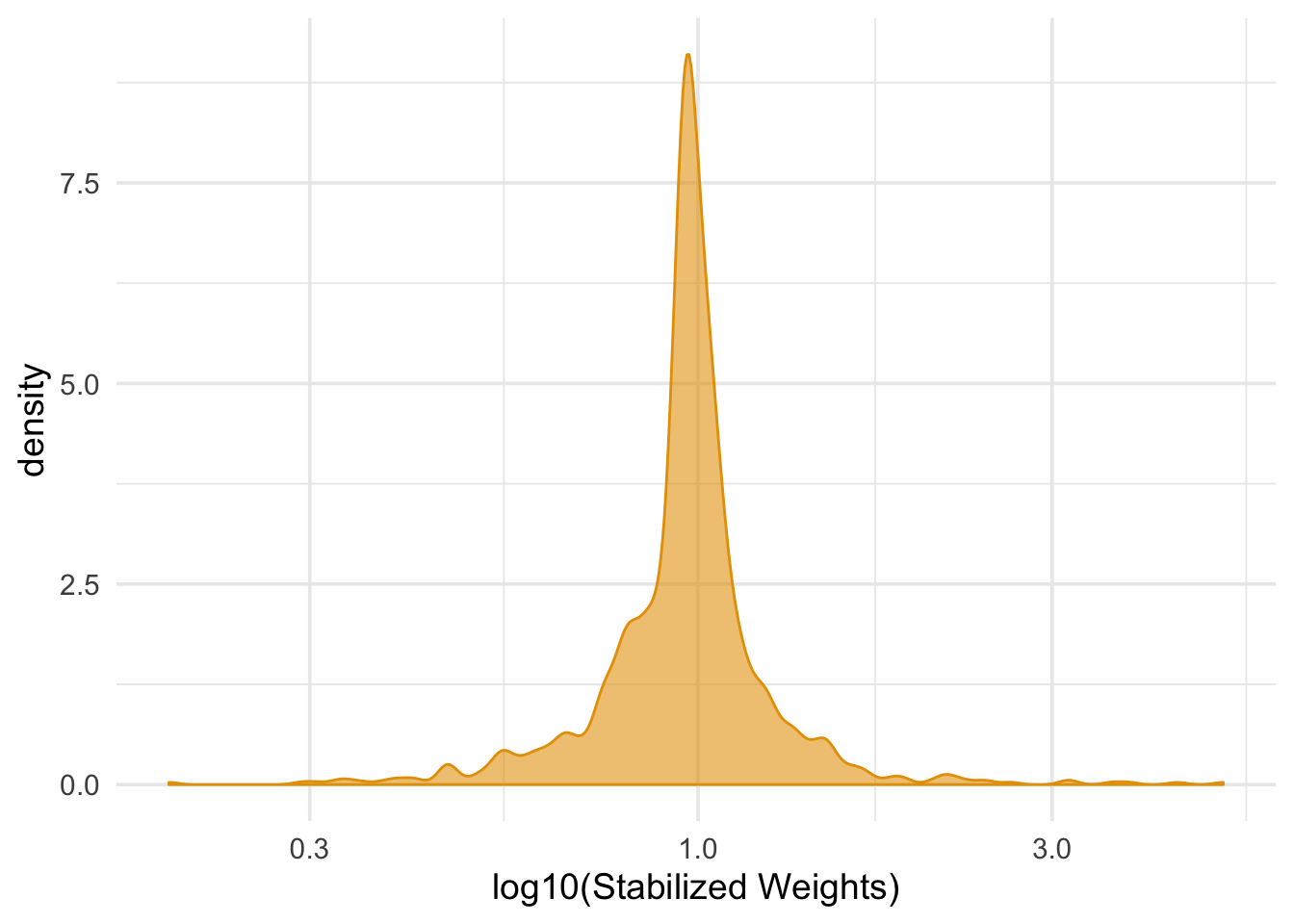
smk_intensity_model <- lm_robust(wt82_71 ~ smkintensity82_71 + I(smkintensity82_71^2), data = nhefs_light_smokers, weights = swts)
smk_intensity_model_ols <- lm(wt82_71 ~ smkintensity82_71 + I(smkintensity82_71^2), data = nhefs_light_smokers, weights = swts)
smk_intensity_model %>%
tidy()## term estimate std.error statistic p.value
## 1 (Intercept) 2.004524173 0.304377051 6.585661 6.851793e-11
## 2 smkintensity82_71 -0.108988851 0.032772315 -3.325638 9.097994e-04
## 3 I(smkintensity82_71^2) 0.002694946 0.002625509 1.026447 3.048951e-01
## conf.low conf.high df outcome
## 1 1.407332469 2.601715878 1159 wt82_71
## 2 -0.173288557 -0.044689144 1159 wt82_71
## 3 -0.002456336 0.007846227 1159 wt82_71calculate_contrast <- function(.coefs, x) {
.coefs[1] + .coefs[2] * x + .coefs[3] * x^2
}
boot_contrasts <- function(data, indices) {
.df <- data[indices, ]
coefs <- lm_robust(wt82_71 ~ smkintensity82_71 + I(smkintensity82_71^2), data = .df, weights = swts) %>%
tidy() %>%
pull(estimate)
c(calculate_contrast(coefs, 0), calculate_contrast(coefs, 20))
}
bootstrap_contrasts <- nhefs_light_smokers %>%
boot(boot_contrasts, R = 2000)
bootstrap_contrasts %>%
tidy(conf.int = TRUE, conf.meth = "bca")## # A tibble: 2 x 5
## statistic bias std.error conf.low conf.high
## <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2.00 -0.0338 0.300 1.44 2.63
## 2 0.903 0.221 1.38 -1.27 3.882.5 Program 12.5
logistic_msm <- geeglm(
death ~ qsmk,
data = nhefs_complete,
family = binomial(),
std.err = "san.se",
weights = swts,
id = id,
corstr = "independence"
)
tidy(logistic_msm, conf.int = TRUE, exponentiate = TRUE) ## # A tibble: 2 x 7
## term estimate std.error statistic p.value conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0.225 0.0789 356. 0 0.193 0.263
## 2 qsmk 1.03 0.157 0.0367 0.848 0.757 1.402.6 Program 12.6
numerator_sex <- glm(qsmk ~ sex, data = nhefs_complete, family = binomial())
nhefs_complete <- numerator_sex %>%
augment(type.predict = "response", data = nhefs_complete %>% select(-.fitted:-.std.resid)) %>%
mutate(numerator_sex = ifelse(qsmk == 0, 1 - .fitted, .fitted)) %>%
select(id, numerator_sex) %>%
left_join(nhefs_complete, by = "id") %>%
mutate(swts_sex = numerator_sex * wts)
nhefs_complete %>%
summarize(mean_wt = mean(swts_sex), sd_wts = sd(swts_sex))## # A tibble: 1 x 2
## mean_wt sd_wts
## <dbl> <dbl>
## 1 0.999 0.271ggplot(nhefs_complete, aes(swts_sex)) +
geom_density(col = "#E69F00", fill = "#E69F0095") +
scale_x_log10() +
theme_minimal(base_size = 14) +
xlab("log10(Stabilized Weights)")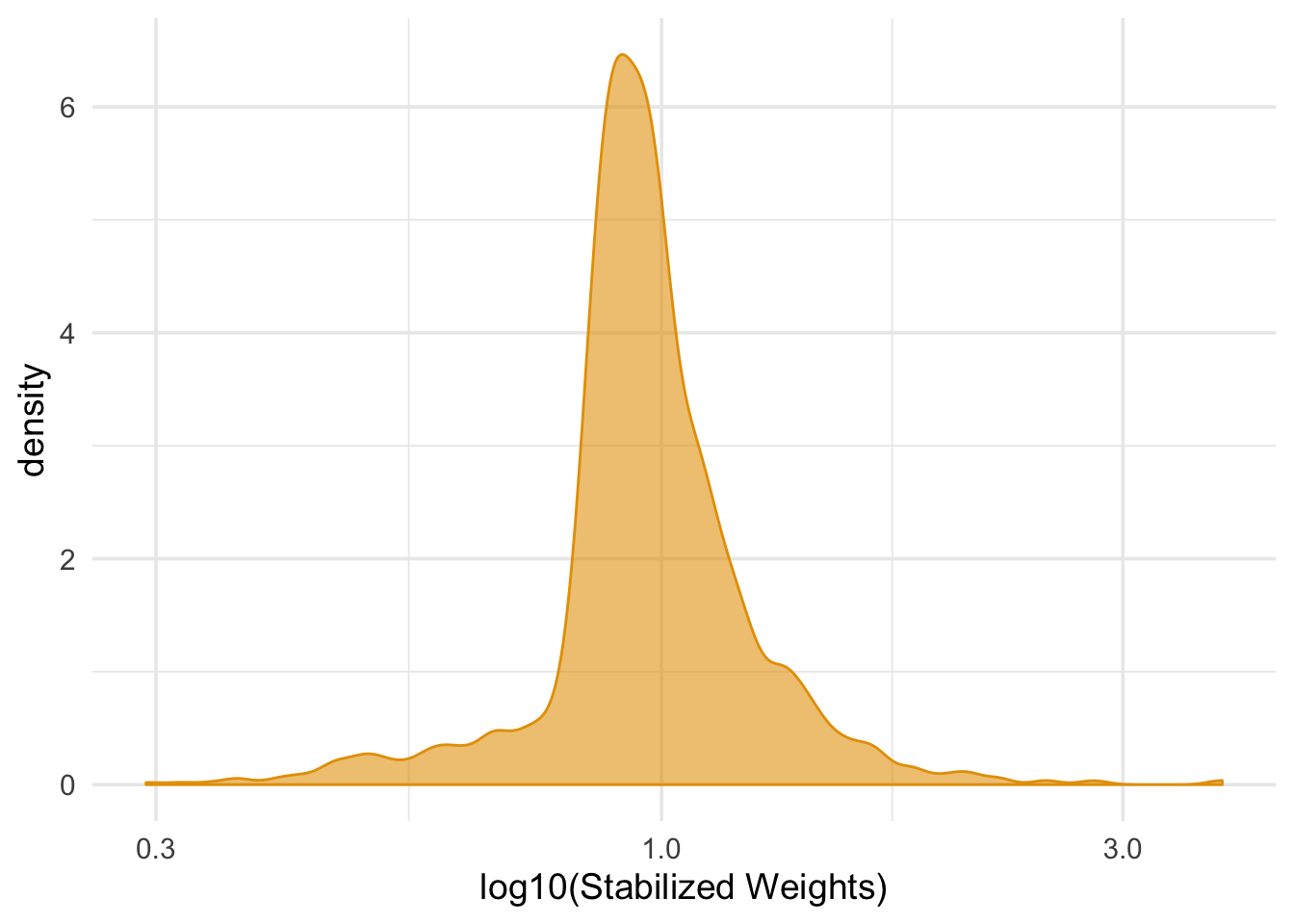
## term estimate std.error statistic p.value conf.low
## 1 (Intercept) 1.784446876 0.3101597 5.75331559 1.051124e-08 1.1760735
## 2 qsmk 3.521977634 0.6585705 5.34791301 1.021696e-07 2.2302023
## 3 sex1 -0.008724784 0.4492401 -0.01942121 9.845076e-01 -0.8899019
## 4 qsmk:sex1 -0.159478525 1.0498718 -0.15190286 8.792832e-01 -2.2187851
## conf.high df outcome
## 1 2.3928202 1562 wt82_71
## 2 4.8137530 1562 wt82_71
## 3 0.8724524 1562 wt82_71
## 4 1.8998280 1562 wt82_712.7 Program 12.7
# using complete data set
nhefs_censored <- nhefs %>%
drop_na(qsmk, sex, race, age, school, smokeintensity, smokeyrs, exercise,
active, wt71)
numerator_sws_model <- glm(qsmk ~ 1, data = nhefs_censored, family = binomial())
numerators_sws <- numerator_sws_model %>%
augment(type.predict = "response", data = nhefs_censored) %>%
mutate(numerator_sw = ifelse(qsmk == 0, 1 - .fitted, .fitted)) %>%
select(id, numerator_sw)
denominator_sws_model <- glm(
qsmk ~ sex + race + age + I(age^2) + education +
smokeintensity + I(smokeintensity^2) +
smokeyrs + I(smokeyrs^2) + exercise + active +
wt71 + I(wt71^2),
data = nhefs_censored, family = binomial()
)
denominators_sws <- denominator_sws_model %>%
augment(type.predict = "response", data = nhefs_censored) %>%
mutate(denominator_sw = ifelse(qsmk == 0, 1 - .fitted, .fitted)) %>%
select(id, denominator_sw)
numerator_cens_model <- glm(censored ~ qsmk, data = nhefs_censored, family = binomial())
numerators_cens <- numerator_cens_model %>%
augment(type.predict = "response", data = nhefs_censored) %>%
mutate(numerator_cens = ifelse(censored == 0, 1 - .fitted, 1)) %>%
select(id, numerator_cens)
denominator_cens_model <- glm(
censored ~ qsmk + sex + race + age + I(age^2) + education +
smokeintensity + I(smokeintensity^2) +
smokeyrs + I(smokeyrs^2) + exercise + active +
wt71 + I(wt71^2),
data = nhefs_censored, family = binomial()
)
denominators_cens <- denominator_cens_model %>%
augment(type.predict = "response", data = nhefs_censored) %>%
mutate(denominator_cens = ifelse(censored == 0, 1 - .fitted, 1)) %>%
select(id, denominator_cens)
nhefs_censored_wts <- nhefs_censored %>%
left_join(numerators_sws, by = "id") %>%
left_join(denominators_sws, by = "id") %>%
left_join(numerators_cens, by = "id") %>%
left_join(denominators_cens, by = "id") %>%
mutate(
swts = numerator_sw / denominator_sw,
cens_wts = numerator_cens / denominator_cens,
wts = swts * cens_wts
)
nhefs_censored_wts %>%
summarize(mean_wt = mean(cens_wts), sd_wts = sd(cens_wts))## # A tibble: 1 x 2
## mean_wt sd_wts
## <dbl> <dbl>
## 1 0.999 0.0519ggplot(nhefs_censored_wts, aes(cens_wts)) +
geom_density(col = "#E69F00", fill = "#E69F0095") +
scale_x_log10() +
theme_minimal(base_size = 14) +
xlab("log10(Stabilized Weights)")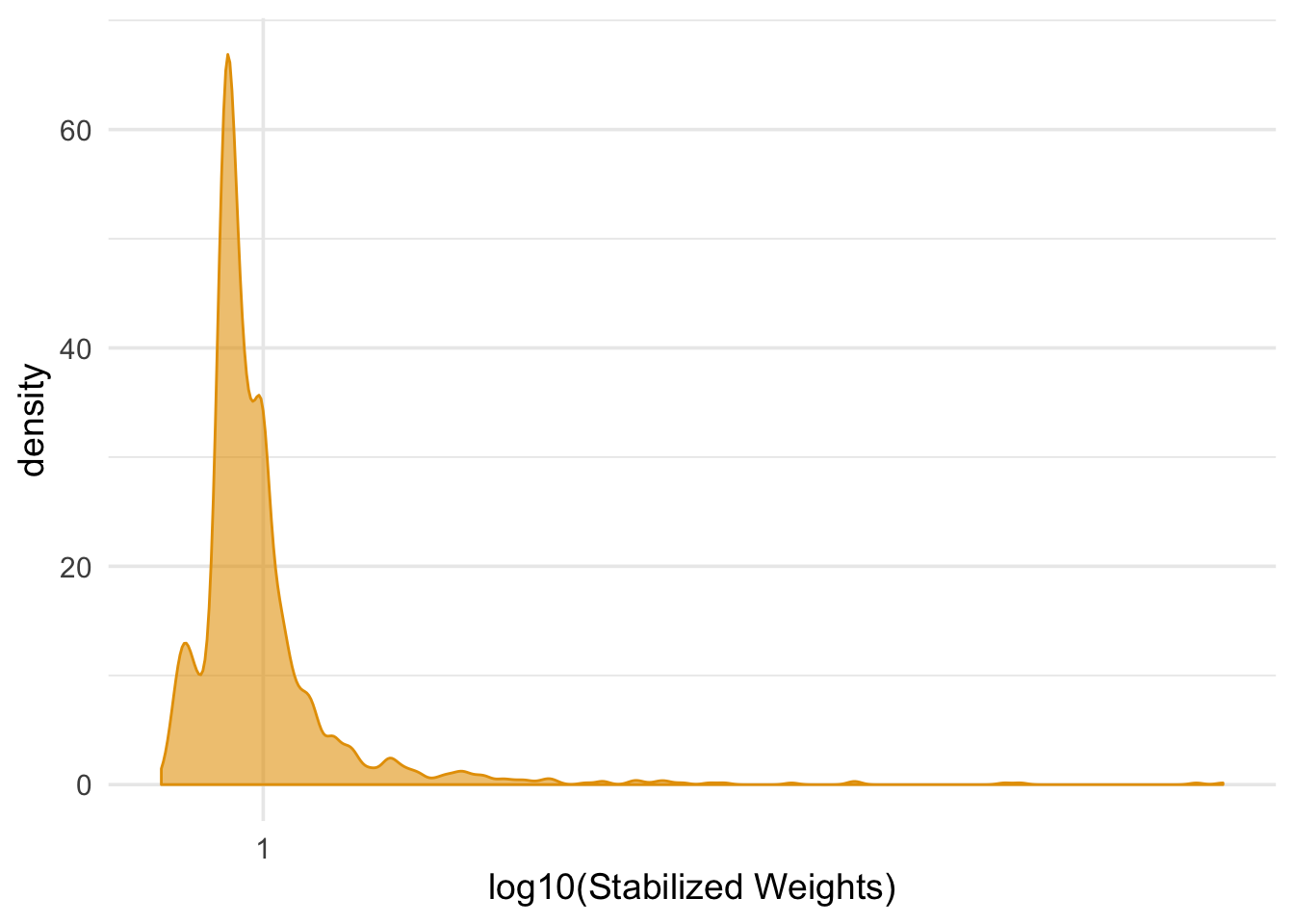
## term estimate std.error statistic p.value conf.low conf.high
## 1 (Intercept) 1.661990 0.2328693 7.137009 1.454358e-12 1.205221 2.118759
## 2 qsmk 3.496493 0.5265564 6.640301 4.305944e-11 2.463662 4.529324
## df outcome
## 1 1564 wt82_71
## 2 1564 wt82_71